
Image: Mattias Pettersson

Image: Mattias Pettersson
Research group Our lab is interested in understanding the molecular mechanism of autophagy and membrane trafficking regulated by small GTPases by developing novel chemical and chemo-optogenetic approaches.
Yaowen Wu Lab concept
ImageYaowen WuThe foundation of living systems isbased on chemical reactions. They are spatiotemporally regulated in response to extra- or intracellular stimuli. Of importance to understand the mechanism of life is the ability to visualize and to perturb these reactions. Chemical tools provide unique means to emulate or modulate biological systems to either investigate the underlying biology or create new function.
In my laboratory, we run both biology-driven and chemistry-driven projects. In the former, we have defined biological questions, which are not readily solved by traditional biological methods. We tackle these problems using novel chemical approaches in combination with biochemical and cell biological methods. In the latter, we develop novel chemical tools that can be widely used to investigate biology. These approaches make it possible to reveal new mechanism of biology.
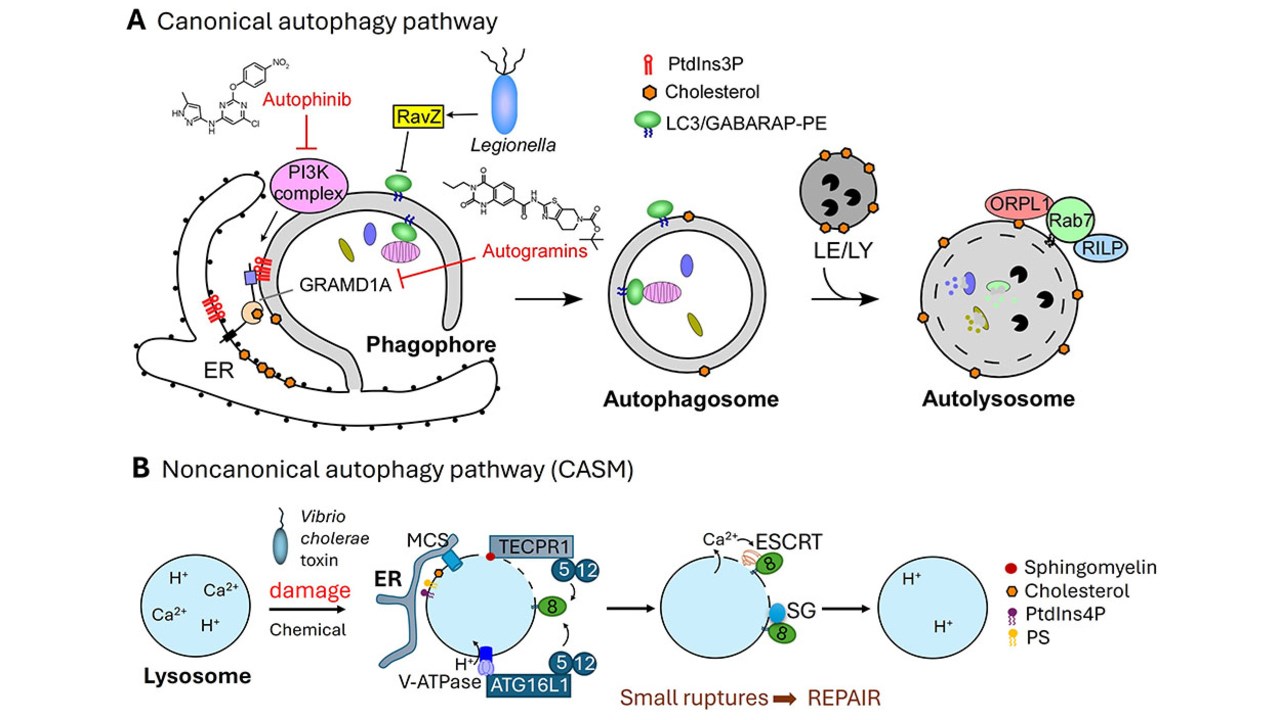
Autophagy is an evolutionarily conserved self-eating process in eukaryotes, mainly involved in elimination of organelles and aggregated proteins. Malfunction of autophagy has been associated with diverse human diseases, including cancer, neurodegeneration, cardiac hypertrophy, and pathogen infection. Formation of double-membrane autophagosome is the key process in autophagy. The regulatory pathways of autophagy and the formation of autophagosome remain the most important questions for understanding autophagy.
Using a combination of cell biology, structural biology and chemical genetic approaches, we have elucidated fundamental mechanisms underlying autophagy. Our laboratory has elaborated a novel mode of action for virulent bacteria (Legionella pneumophila) against host autophagy (eLife 2017, Virulence 2019 review, ChemBioChem 2020). We have established that Rab33B plays an essential role in autophagosome formation by targeting the E3-like ATG16L1-ATG5–ATG12 complex to the forming autophagosome (Autophgay 2022). We have identified novel chemotypes for autophagy modulation and new cellular targets involved in new mechanism of autophagy regulation, e.g. the GRAMD1A-cholesterol axis in autophagosome biogenesis (Angew Chem 2017, Angew Chem 2017, Chem Sci 2018, MiMB 2018, BMC 2019, Nat Chem Biol 2019, Angew Chem 2020, Angew Chem 2020, Cell Chem Biol 2021).
Our lab has gained new insights into noncanonical autophagy (or Conjugation of ATG8 to Single Membranes, CASM). We identify novel compounds (e.g. pseudo-natural products) that induce CASM and characterize the mechanisms of the Vibrio cholerae cytotoxin MakA inducing CASM (JCS 2021, JCB 2022, Autophagy 2022, Angew Chem 2022, ChemBioChem 2024, PNAS 2024). We and others discover the sphingomyelin-TECPR1 axis and TECPR1-ATG5–ATG12 as a new E3-like ligase for ATG8 conjugation (STIL pathway) in lysosomal membrane repair, and elucidated molecular mechanisms for noncanonical functions of autophagy proteins (EMBO Rep 2023, Autophagy 2024, bioRxiv 2024).
ImageYaowen Wu
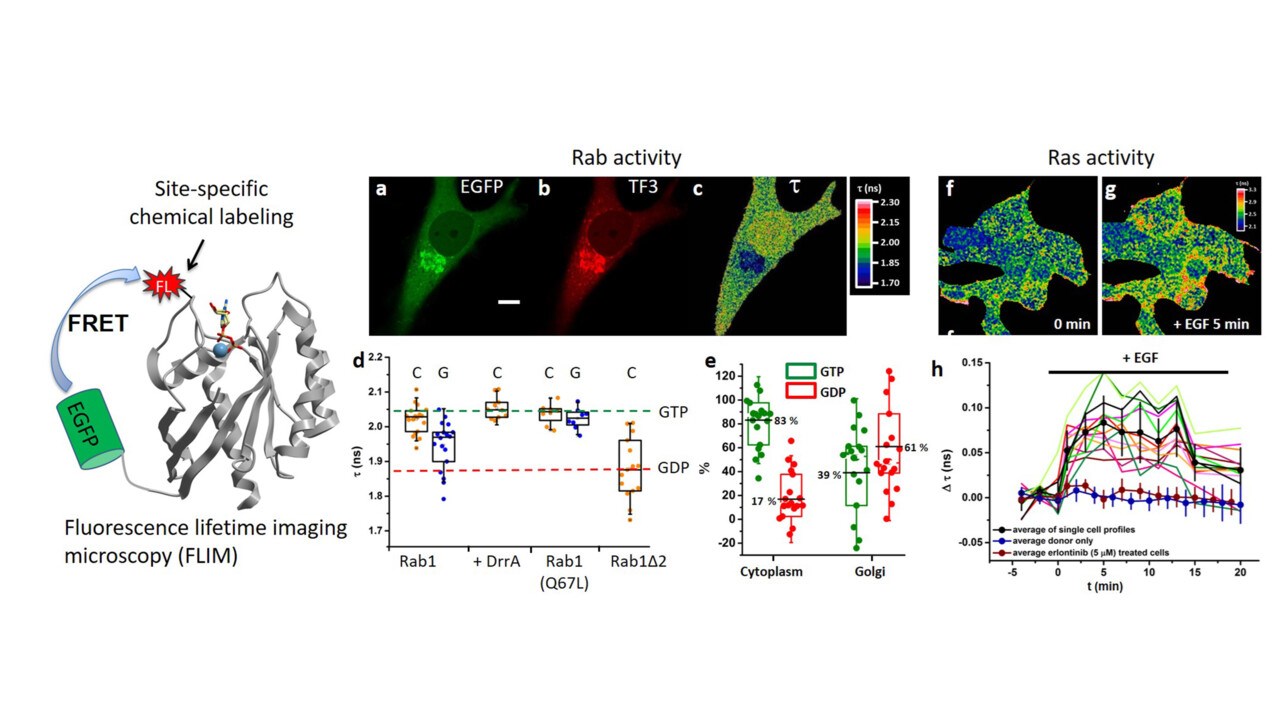
Due to the spatial segregation of intracellular organelles of eukaryotic cells a sophisticated system of vesicular transport is required to ensure the communication between different compartments. Rab GTPases function as key regulators of intracellular vesicular transport. With the help a complex network of Rab regulators and effectors, Rab GTPases regulate these processes through tightly controlled enzymatic GTPase cycle and spatial distribution in cells. We are interested in answering the question of how these complex biological reactions are coordinated and how the malfunction of these processes link to diseases. These questions have been assessed at both the molecular and the cellular level in a quantitative manner. We established a comprehensive new model for membrane targeting of Rab GTPases, the key organizers of membrane trafficking (PNAS 2014, PNAS 2016, Biochemistry 2019, MiMB 2021, MiMB 2021).
ImageYaowen Wu
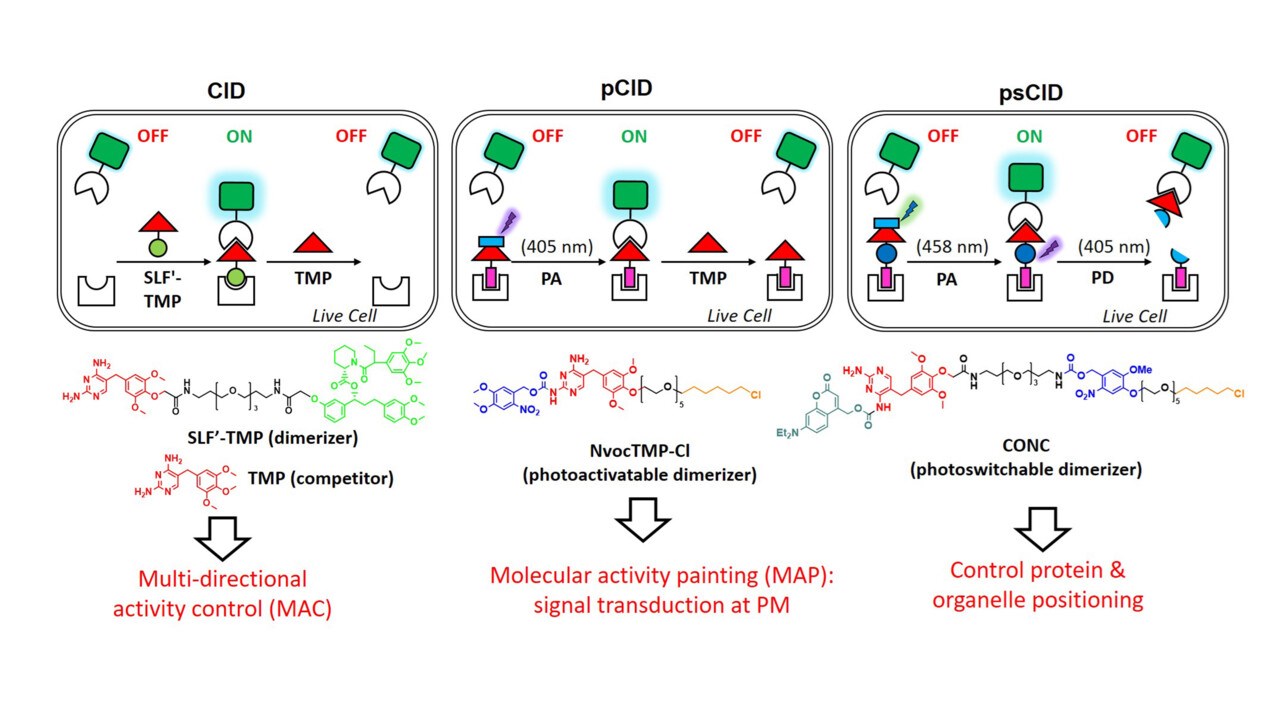
Genetic perturbations by gain or loss of gene function through overexpression, knock-out or knock-down approaches are powerful for biological studies. However, traditional genetic approaches are chronic (hours to days). Consequently, the phenotype may not be detected due to adaptation and the dynamics of phenotypic change cannot be followed. Chemical/light-inducible dimerization approaches provide means to control biological systems with spatial and temporal resolution that is unmatched by traditional genetic perturbations (Curr Opin Chem Biol 2015, Chemistry 2019, Nat Meth 2023, reviews). They have been very useful to dissect the complexity of biological regulatory networks (JACS 2012).
Our laboratory has developed the first bioorthogonal and reversible chemically induced dimerization (CID) system (SLF'-TMP system) for controlling protein function by small molecules in live cells (Angew Chem 2014). A photoactivatable CID (pCID) and photoactivatable/photocleavable CID system (psCID) using photo-caged dimerizer (NovcTMP-Cl) (Angew Chem 2017) and photo-switchable dimerizer (CONC) allow superior spatiotemporal control of protein function in live cells (Angew Chem 2018). We have developed novel "Molecular Activity Painting (MAP)" strategy by combining pCID with immobilized artificial receptors to enable "painting" of signaling molecules and their activity at micrometer-scale at the plasma membrane. Using photoactivatable dual-chemically induced dimerization (pdCID) system, we developed a Multi-directional Activity Control (MAC) approach to spatiotemporally control cellular signaling and intracellular cargo transport (Angew Chem 2018).
ImageYaowen Wu
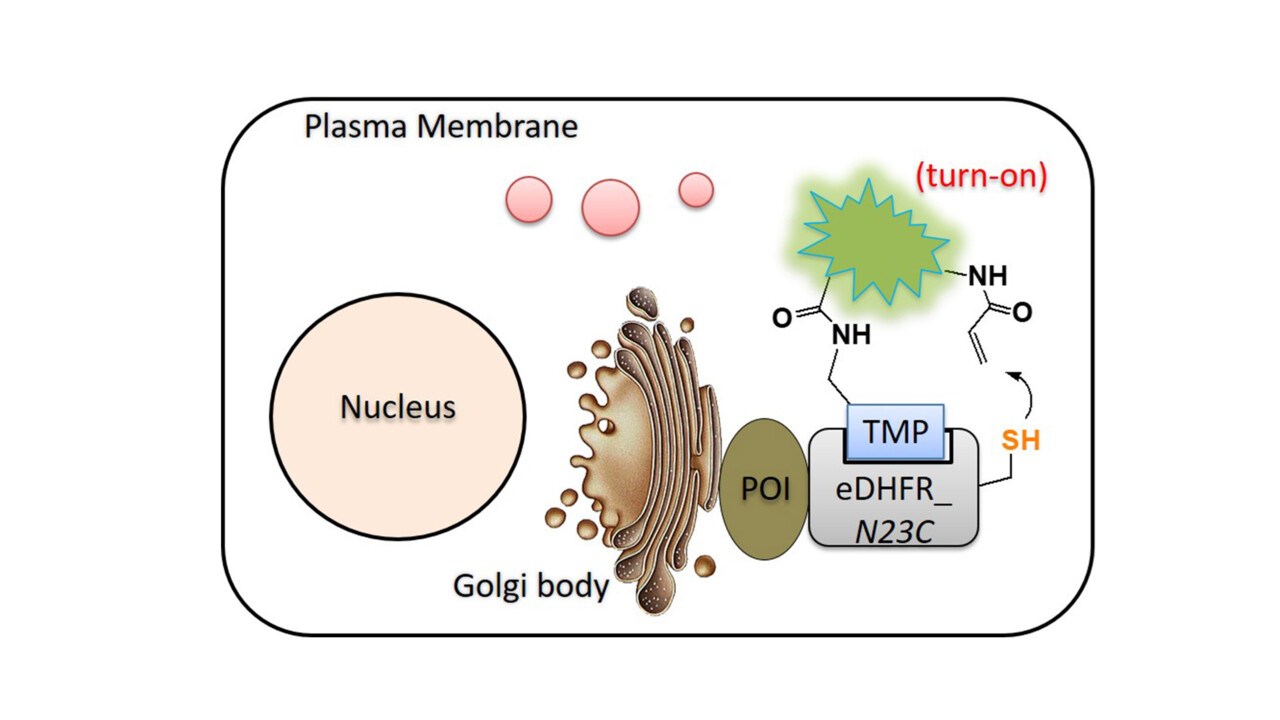
Labeling proteins with synthetic probes, such as fluorophores, affinity tags, and other functional labels is enormously useful for characterizing protein function in vitro, in live cells, or in whole organisms. Chemical methods have substantially expanded the tools that are can be used to modify proteins. Our work has been focused on preparation of post-translationally modified proteins (ChemBioChem 2012, 2013, 2020, Topics Curr Chem 2015 review, Bioorg Med Chem 2017, MiMB 2018). These semi-synthetic protein probes have been very useful in the investigation of protein functions in membrane trafficking and autophagy. We are interested in developing new methods for protein modification/labeling (Angew Chem 2011, JPS 2014, JACS 2014, Chem Comm 2015, Org Bioorg Chem 2016 review, Angew Chem 2016, MiMB 2019, Chem Sci 2022). These methodologies have been valuable for visualizing biochemical reactions in living systems and proteomic studies. Such developments not only facilitate the study of the system we are interested but also contribute new tools to the larger biological field.
ImageYaowen Wu
Yaowen Wu received his BS in Chemistry from Sun Yat-sen University in 2001 and his MS in Organic Chemistry from Tsinghua University in 2004 in China. After graduating as Dr rer. nat. (2008) at the Technische Universität Dortmund working at the Max Planck Institute of Molecular Physiology in Germany and a postdoctoral study in cell biology at King’s College London, he has been leader of an Otto Hahn group at the Max Planck Institute in Dortmund since 2010. Since 2012, he has been group leader of Chemical Genomics Centre of the Max Planck Society. He was then appointed as Professor in Biochemistry at the Umeå University in 2018. He has served as Director of Umeå Centre for Microbial Research (UCMR) since 2020.
His research group develops small molecules and new chemical methods to modify proteins or manipulate protein function in the context of biological systems with a particular focus on regulatory mechanisms in membrane trafficking and autophagy. He has received awards and honors including Göran Gustafsson Prize in Molecular Biology “for his innovative molecular studies of intracellular transport and autophagy” (awarded by Royal Swedish Academy of Sciences annually to one outstanding scientist of < 45 years old in each field of mathematics, physics, chemistry, molecular biology and medicine), "Future of Biochemistry" recognized by “tackling problem that transcend traditional field boundaries and challenges” in the field of biochemistry, Wallenberg Academy Fellow (Royal Swedish Academy of Sciences), European Research Council (ERC) Investigator, Behrens-Weise Award, Biomedicine Research Prize and Otto-Hahn Award (selected as the single awardee of the Otto-Hahn Group Leader in 2009 in the biomedical section of the Max Planck Society).
Knyazeva A, Li S, Corkery D, Shankar K, Herzog L, Zhang X, Singh B, Niggemeyer G, Grill D, Gilthorpe J, Gaetani M, Carlson LA, Waldmann H, Wu YW*. (2024) A chemical inhibitor of IST1-CHMP1B interaction impairs endosomal recycling and induces noncanonical LC3 lipidation. Proc. Natl. Acad. Sci. U. S. A. 121 (17), e2317680121.
Corkery DP, Castro-Gonzalez S, Knyazeva A, Herzog LK, Wu YW*. (2023) An ATG12-ATG5-TECPR1 E3-like complex regulates unconventional LC3 lipidation at damaged lysosomes. EMBO Rep: e56841.
Comment in Florey O. TECPR1 helps bridge the CASM during lysosome damage. EMBO J. 2023: e115210; Comment in Deretic V, Klionsky, DJ. An expanding repertoire of E3 ligases in membrane Atg8ylation. Nat Cell Biol. 2024, 26: 307–308.
Laraia L, Friese A, Corkery DP, Konstantinidis G, Erwin N, Hofer W, Karatas H, Klewer L, Brockmeyer A, Metz M, Schölermann B, Dwivedi M, Li L, Rios-Munoz P, Köhn M, Winter R, Vetter IR, Ziegler S, Janning P, Wu YW*, Waldmann H*. (2019) The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis. Nat. Chem. Biol. 15(7):710-720.
Comment in Aldrich LN. Lipids lead the way. Nat. Chem. Biol. 2019, 15(7):653-654
Chen X, Wu YW*. (2018) Tunable and photoswitchable chemically induced dimerization for chemo-optogenetic control of protein and organelle positioning. Angew. Chem. Int. Ed. 57 (23): 6796-6799.
Yang A, Pantoom S, Wu YW*. (2017) Elucidation of anti-autophagy mechanism of the Legionella effector RavZ using semisynthetic LC3 proteins. eLife. pii: e23905.
Chen X, Venkatachalapathy M, Kamps D, Weigel S, Kumar R, Orlich M, Garrecht R, Hirtz M, Niemeyer CM, Wu YW*, Dehmelt L*. (2017) “Molecular Activity Painting”: Switch-like, light-controlled perturbations inside living cells. Angew. Chem. Int. Ed. 56 (21):5916-5920 (Hot paper, inside cover story)
Voss S, Krüger DM, Koch O, Wu YW*. (2016) Spatiotemporal imaging of small GTPases activity in live cells. Proc. Natl. Acad. Sci. U. S. A. 113 (50):14348-14353.
Liu P, Calderon A, Konstantinidis G, Hou J, Voss S, Chen X, Li F, Banerjee S, Hoffmann JE, Theiss C, Dehmelt L, Wu YW*. (2014) A bioorthogonal small-molecule switch system for controlling protein function in live cells. Angew. Chem. Int. Ed. 53 (38):10049-55.
Li F, Yi L, Zhao L, Itzen A, Goody RS, Wu YW*. (2014) The role of the hypervariable C-terminal domain in Rab membrane targeting. Proc. Natl. Acad. Sci. U. S. A. 111 (7): 2572-7. Recommended by Faculty1000
Liu W, Li F, Chen X, Hou J, Yi L, Wu YW*. (2014) A rapid and fluorogenic TMP-AcBOPDIPY probe for covalent labeling of proteins in live cells. J. Am. Chem. Soc. 136 (12): 4468-71.

Professor Yaowen Wu's lab is at the forefront of developing chemo-optogenetic systems.

PNAS study makes a good "case" for using small molecules as chemical tools to understand complex biology.

The great interest highlights the need for meeting platforms focusing specifically on postdocs and PhDs.